[su_dropcap] It[/su_dropcap] is common knowledge that genetic information of organisms is encoded in DNA in the form of sequential interactions of merely four nucleotide bases, Adenine(A), Thymine(T), Guanine(G), and Cytocine(C). This seemingly simple yet elegant design of genetic coding manifests one prime example of nature’s mystique and efficiency; It is this design that gives rise to the amino acids (and proteins), which are responsible for the proper functioning of life activities. Yet, chemists today are attempting to challenge nature’s reign over genetic encoding. Installing artificial base pairs is one such approach.
Research on artificial base pairs is founded on the premise first asserted in 1962 by Alexander Rich as shown below;
[su_quote]the amount of information stored in DNA may be increased if the number of base pairs could be expanded[/su_quote]
For example the number of codons, or triplets of base pairs used to produce a single amino acid, may be increased from 64 (4 x 4 x4) to 216 (6 x 6 x 6) by simply installing a 5th and 6th artificial nucleotide base. Applications in artificial protein synthesis could be envisioned, in turn, if the increased codons could each yield a specific amino acid outside the universal 20. Plenty other applications are imaginable. New functions of DNA and RNA themselves, which neither relate to information storage nor replication, are conceivable.
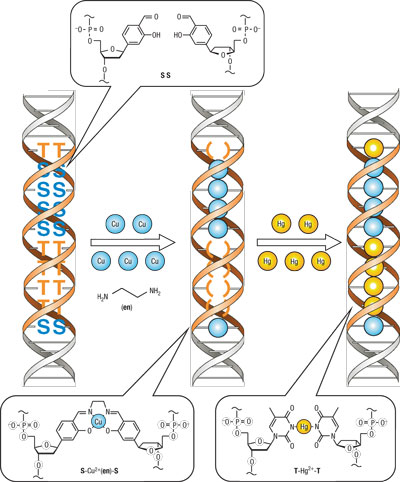
In fact, numerous examples and applications have been reported on installing artificial base pairs through organic synthesis. In one such case, Prof. Shionoya and his research group at the University of Tokyo have used artificial base pairs in DNA as ligands of metal complexes to develop a method to self assemble metal ions within the double helices.[1] This highly creative example illustrates how artificial DNA could be applied to novel nanomaterial synthesis.
Contrary to belief, finding combinations of artificial base pairs is not an insurmountable task. Finding a proper combination with a sufficiently high replication accuracy, however, may be. In nature, DNA is replicated by an enzyme called DNA polymerase. Error-rates in DNA polymerase are on the order of 1 in 104 (proof-reading processes further increase this number to 1 in 109!). Such extreme selectivity is rarely observed in artificial systems, and has previously been believed to be attainable only through extreme trial and error repetitions on structural fine-tuning.
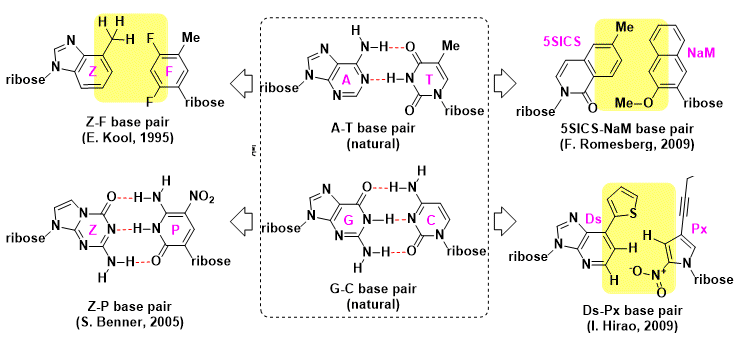
Previous structural analysis on A-T and C-G pair interactions have lead to some insights on properties necessary for high accuracy replication. These insights have shown that base pair geometry, dipole moment, and stacking are some important characteristics; hydrogen-bonding is not a requisite. Shown below are some examples of base pairs that have been replicated through PCR (polymerase chain reaction).
To the delight of the community, a breakthrough in this field has been recently reported. Through years of effort that amounts to a colossal achievement, an artificial base pair [7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) and 2-nitro-4-propynylpyrrole (Px)] that is shown to be replicable by DNA polymerase (Ds-Px: 99.9%/cycle) has been developed! An application of this example will be discussed in a subsequent post.
This post is an English translation of the original blog post written in Japanese. The original post can be found here.
-
Reference
[1] “Programmable self-assembly of metal ions inside artificial DNA duplexes” Shionoya, M. et al. Nat. Nanotech. 2006, 1, 190. doi:10.1038/nnano.2006.141
-
Related Book
[amazonjs asin=”0849314267″ locale=”US” title=”Artificial DNA: Methods and Applications”][amazonjs asin=”162703109X” locale=”US” title=”Biolistic DNA Delivery: Methods and Protocols (Methods in Molecular Biology)”]
-
Related Links
Shionoya Laboratory, University of Tokyo