Zirkin, S.; Fishman, S.; Sharim, H.; Michaeli, Y.; Don, J.; Ebenstein, Y. J. Am. Chem. Soc., 2014, 136, 7771–7776.
DOI: 10.1021/ja503677n
DNA damage and repair are linked to fundamentalbiological processes such as metabolism, disease, and aging. Single-strand lesions are the most abundant form of DNA damage; however,methods for characterizing these damage lesions are lacking. To avoiddouble-strand breaks and genomic instability, DNA damage isconstantly repaired by efficient enzymatic machinery. We take advantageof this natural process and harness the repair capacity of a bacterialenzymatic cocktail to repair damaged DNA in vitro and incorporatefluorescent nucleotides into damage sites as part of the repair process.We use single-molecule imaging to detect individual damage sites ingenomic DNA samples. When the labeled DNA is extended on amicroscope slide, damage sites are visualized as fluorescent spots alongthe DNA contour, and the extent of damage is easily quantified. Wedemonstrate the ability to quantitatively follow the damage dose response to different damaging agents as well as repair dynamics in response to UV irradiation in several cell types. Finally, we show the modularity of this single-molecule approach by labeling DNA damage in conjunction with 5-hydroxymethylcytosine in genomic DNA extracted from mouse brain tissue.
Introduction
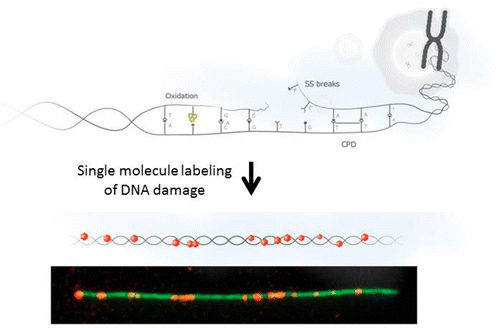
DNA is constantly damaged by various external stimuli like UV radiation, cosmic ray and ROS (reactive oxygen species) and so forth. Since the damaged DNA often leads to many illnesses, to further understand its mechanism is very important. Yuval Ebenstein at Raymond and Beverly Sackler Faculty of Exact Sciences challenged this by simply making the damaged DNA base pair visible with conventional microscopy.
Method
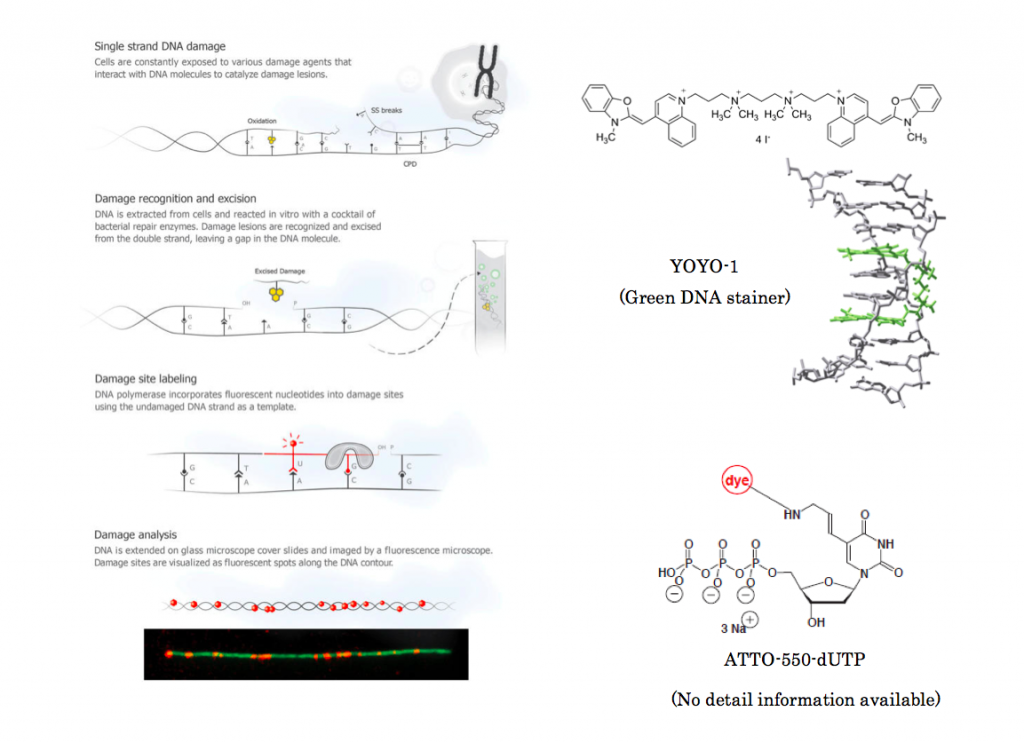
The author first extended single stranded DNA on glass slide and stained it with YOYO-1 stainer based on their previously reported procedure.[1] Then ATTO-550-dUTP and other three corresponding base constituents were added followed by either UV radiation or further addition of ROS to deliberately damage DNA base pair. Then the damage base pair is repaired through DNA polymerase,[2] making the damaged bases to be labeled with red fluorescent dots.
Observation of damaged DNA.
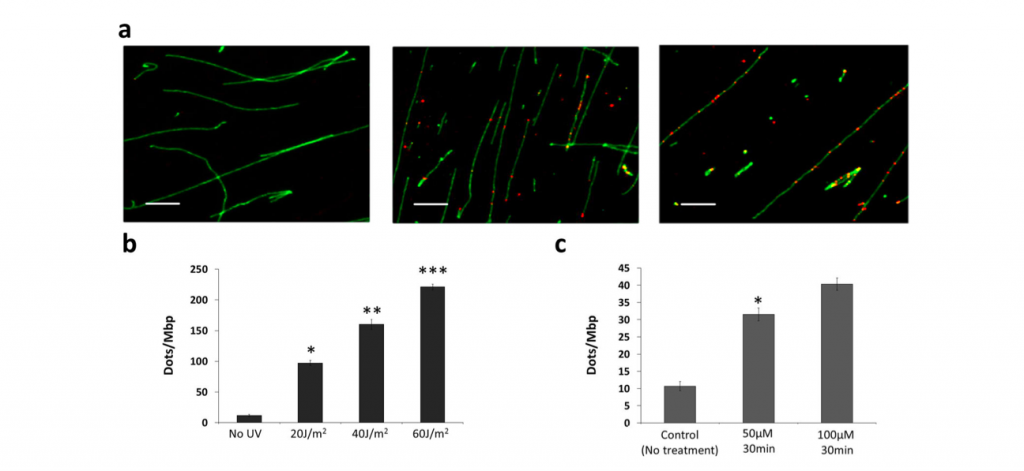
The extended DNA on the glass plate were exposed to UV (with intensity from 20-60 J/m2) and ROS reagents (50 or 100 μM) respectively to analyze the change in the red fluorescent spot. Expectedly, the number of red spot, the repaired bases position, increased in proportion to that of DNA damager.
Reference
[1]”Optical detection of epigenetic marks: sensitive quantification and direct imaging of individual hydroxymethylcytosine bases”
Michaeli, Y.; Shahal, T.; Torchinsky, D.; Grunwald, A.; Hoch, R.; Ebenstein, Y. Chem. Commun. 2013, 49, 8599.
DOI: 10.1039/C3CC42543F
[2]”DNA damage and repair”