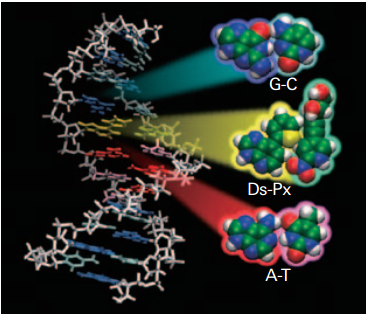
In a previous post, we discussed the myriad applications of artificial nucleotide base pairs. In this post, I’d like to highlight an exquisite example that was demonstrated by Hirao and coworkers at RIKEN in 2013.[1] This recent report introduced an artificial DNA aptamer that, astonishingly, exhibited an affinity greater than that observed in natural nucleotides.
-
The key to success lies in high replication accuracy
Hirao and coworkers have previously reported on the Px-Ds pair, which was the first example of an hydrophobic nucleotide base pair.[2] This pair exhibited a remarkable replication accuracy of over 99.9% per cycle.
The path to this seminal work was far from simple, however. Hirao originally began this work in 1997, attempting numerous optimizations in the form of functional groups with various steric influences, adding/subtracting hydrogen bonds, examining lock and key-esque shape fits, and dedicating 10 years of delicate research.
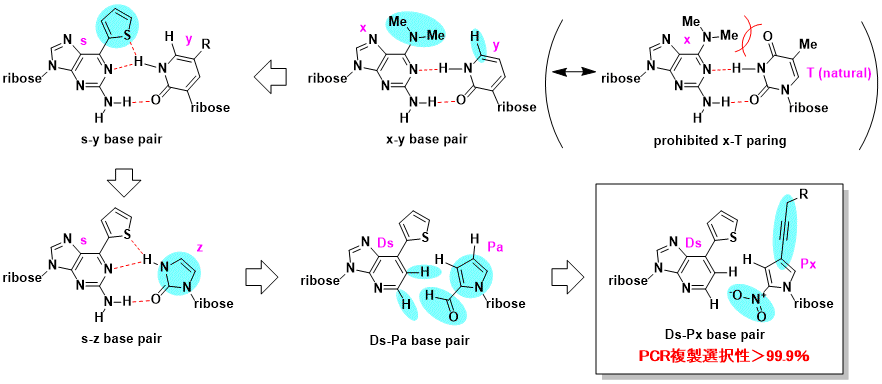 The path to perfection. Taken from ref [2].
The path to perfection. Taken from ref [2].
-
Installing a Ds base in a DNA aptamer.
DNA aptamers are used in nucleic acid medication by taking advantage of the affinity of an oligonucleotide towards a specific protein. In essence, the medication is an analogue of antibodies. In contrast to the use of antibodies, which requires an immune response for its synthesis, DNA aptamers possess the advantage of stability, low cost, short lead time, and mass production, granted that the base pair sequence is known.
On the other hand, the existence of only four nucleotides has placed limitations with respect to structure variation, a key aspect that antibodies take advantage of. The work highlighted in this post overcomes this obstacle in the form of artificial base pairs.
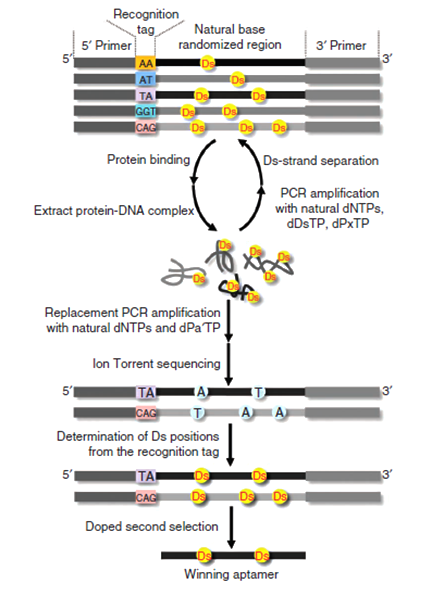
Hirao and coworkers prepared a DNA library (around 40 base paris, approximately 1.8×1014 combinations), whereby each component contains 1~3 Ds bases. After seven steps of SELEX, a combi-chem method of screening oligonucleotides, Hirao discovered an aptamer with a strong affinity for VEGF-165, a signal protein associated with age-related muscular degeneration (AMD).
Impressively, the aptamers that Hirao discovered possess a Kd of up to 0.65pM, which is 100 times greater than Macugen (pegaptanib), a nucleic acid drug that is approved for AMD treatment in Japan. The optimal aptamer contains three Ds bases, and studies have shown that replacing these with Adenine results in significant loss in affinity; the effect of installing Ds bases has thereby been demonstrated.
-
Prospects
Whereas DNA aptamers have so far struggled in medicinal applications, Hirao’s method of installing hydrophobic artificial base pairs has opened up new possibilities. Hirao’s contributions are further amplified by demonstrating that aptamers with artificial bases could exhibit binding affinities that surpass those of aptamers prepared from natural base pairs.
What must not be forgotten, however, is the countless hours that have been put into this example. Simply discovering an artificial base pair that exhibits high performance and replicability has taken an amount of time comparable to screening for traditional small molecule drugs. It is evident that a task remains in developing a better screening method.
Once discovered, however, an artificial base pair may subsequently lead to a plethora of new results, as shown by the relative short time of finding a VEGF-165 aptamer in comparison to uncovering the Ds-Px pair. This area of research is therefore a highly promising field with prospects of tremendous output.
-
A product of a Galapagos syndrome?
Hirao’s comments, as shown below, manifest an interesting relationship between how research is conducted and its results.
“I have heard through US competitors that a problem they face is that experienced postdocs come and go after only 2~3 years. Research on artificial base pairs requires expertise in organic synthesis as well as molecular biology; knowledge, skill, and instincts that are difficult to acquire in this time span. Our research lab has brought together and maintained a team of experts in these fields for longer periods. I believe that is one of our strengths.”[taken (and translated) from ref [3]]
In contrast to Japan, it is common in the US for researchers to move from one research institute to another, often within several years. Postdocs must rapidly produce results in order to survive the so-called ‘publish or perish’ environment. Rarely does a researcher choose to tackle a project requiring five years or more; a small project with rapid results is preferred. While such a system is desirable for speed and changes, it does not appear best for developing the fundamentals and know-hows.
In this case, the Japanese research system, which comparatively allows for longer periods in nailing the crux of the problem, has worked in its favor. This example seems to demonstrate that maintaining a high flux of researchers is not necessarily desirable; how a research institute is organized also seems to require diversity.
This post is an English translation of the original blog post written in Japanese. The original post can be found here.
-
References
[1] “Generation of high-affinity DNA aptamers using an expanded genetic alphabet” Hirao, I. et al. Nat. Biotech. 2013, 35, 453. doi:10.1038/nbt.2556
[2] 「人工塩基対の分子設計」, 平尾一郎、TCIメール(Japanese Review) [PDF]
[3] 「人工DNAで薬を作る」理研ニュース2013年8月号(Japanese Review) [PDF]
-
Related Books
[amazonjs asin=”B001B3WGAW” locale=”US” title=”Artificial DNA: Methods and Applications”][amazonjs asin=”3319019279″ locale=”US” title=”DNA Computing and Molecular Programming: 19th International Conference, DNA 2013, Tempe, AZ, USA, September 22-27, 2013, Proceedings (Lecture Notes in … Computer Science and General Issues)”]
-
Related Links
Ichiro Hirao Laboratory
TAGCyx Biotechnologies