Roger D. Kornberg (born 1947), biochemist who received the Nobel Prize in Chemistry in 2006 for his research on the molecular basis of eukaryotic transcription. His father, Arthur Kornberg, who won the Nobel Prize in Medicine in 1959 for showing how genetic information is carried between DNA molecules. Roger Kornberg’s brother, Thomas, is also a scientist. He isolated DNA polymerase III used for replication. Kornberg’s other brother is an architect and designs research laboratories.
As he said in an interview:
Science was a part of dinner conversation and an activity in the afternoons and on weekends. Both my parents had fine scientific minds and taught by example how to approach questions and problems in a logical, dispassionate way. Scientific reasoning became second nature. Above all, the joy of science became evident to my brothers and me. (authority:http://www.dnaftb.org)
-
Education and Experiences
1967 B.S. in Chemistry from Harvard University
1972 Ph.D. in Chemistry from Stanford University
1972 Research Associate, Biochemistry, Medical Research Council in Cambridge
1976 Assistant Professor, Harvard Medical School
1978- Professor, Stanford University
-
Awards and Honors
1981 Eli Lilly Award in Biological Chemistry
1982 Passano Award, Passano Foundation
1990 Ciba-Drew Award
1997 Harvey Prize
2001 Welch Prize
2002 ASBMB-Merck Award
2002 Pasarow Award in Cancer Research
2002 Leopold Mayer Prize
2002 Le Grand Prix Charles-Leopold Mayer
2005 Alfred P. Sloan Jr. Prize
2006 Nobel Prize in Chemistry
2006 Louisa Gross Horwitz Prize
-
Research
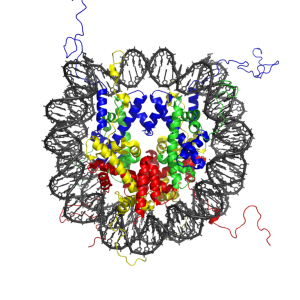
In 1974, Kornberg discovered the nucleosome, the basic unit from which all chromosomes are made.[1]
The organizing principle of the nucleosome, a histoneThe Histones octamer, and its mode of interaction with DNA were deduced from the properties of the tetramer, as follows:
(1) The stoichiometry of the tetramer indicated that apparent deviations from equimolar amounts of H2A, H2B, H3, and H4 in chromatin could be disregarded, and it further implied a repeating unit that contained two each of the histones. The requirement for all four histones to form the repeating unit revealed by X-ray diffraction then led to the proposal of the histone octamer.
(2) Based on the roughly equal weights of histone and DNA in chromatin, an octamer would be associated with about 200 base pairs of DNA.
(3) The biochemical behavior of the tetramer, like that of a typical globular protein, implied a compact shape and thus the wrapping of the DNA on the outside.
(4) The occurrence in chromatin of roughly half as much H1 as each of the other histones pointed to the association of one molecule of H1 with the nucleosome. The lack of a requirement for H1 to reconstitute the X-ray diffraction pattern suggested that H1 bound on the the outside of the nucleosome.
The crystal structure of the nucleosome core particle consisting of H2A , H2B , H3 and H4 core histones, and DNA. (adapted from Wikipedia).
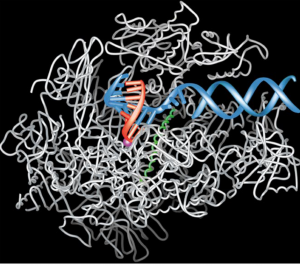
In 2001, Kornberg published the first molecular snapshot of the protein machinery responsible — RNA polymerase — in action.[2]
The transcription process as depicted by Roger Kornberg in 2001. RNA-polymerase in white, DNA-helix in blue and the growing RNA-strand in red (adapted from Nobelprize.org ).
-
References
[1] Kornberg, R.D. Science 1974, 184, 868. 【Pubmed】
[2] a) Cramer, P.; Bushnell D.A.; Kornberg, R.D. Science 2001, 292, 1863. 【Pubmed】;b) Gnatt, A.L.; Cramer, P.; Fu, J.; Bushnell, D.A: Kornberg, R.D. Science 2001, 292, 1876.【Pubmed】
-
Related Links
-
Photo Gallery
-
Movies
-
Related Books
[amazonjs asin=”012391938X” locale=”US” title=”Nucleosomes, Histones & Chromatin Part B, Volume 513 (Methods in Enzymology)”][amazonjs asin=”1244033804″ locale=”US” title=”Nucleosome-ELISA: A novel method to uncover the epiproteomic signatures of mouse and human embryonic stem cells.”]