Akio Arisaka, Rina Mogaki, Kou Okuro, and Takuzo Aida. J. Am. Chem. Soc., 2018, 140, 2687–2692.
DOI: 10.1021/jacs.7b13614
ABSTRACT
We developed dendritic caged molecular glues (CagedGlue-R) as tags for nucleus-targeted drug delivery, whose multiple guanidinium ion (Gu+) pendants are protected by an anionic photocleavable unit (butyrate-substituted nitroveratryloxycarbonyl; BANVOC). Negatively charged CagedGlue-R hardly binds to anionic biomolecules because of their electrostatic repulsion. However, upon exposure of CagedGlue-R to UV light or near-infrared (NIR) light, the BANVOC groups of CagedGlue-R are rapidly detached to yield an uncaged molecular glue (UncagedGlue-R) that carries multiple Gu+ pendants. Because Gu+ forms a salt bridge with PO4–, UncagedGlue-R tightly adheres to anionic biomolecules such as DNA and phospholipids in cell membranes by a multivalent salt-bridge formation. When tagged with CagedGlue-R, guests can be taken up into living cells via endocytosis and hide in endosomes. However, when the CagedGlue-R tag is photochemically uncaged to form UncagedGlue-R, the guests escape from the endosome and migrate into the cytoplasm followed by the cell nucleus. We demonstrated that quantum dots (QDs) tagged with CagedGlue-R can be delivered efficiently to cell nuclei eventually by irradiation with light.
Introduction
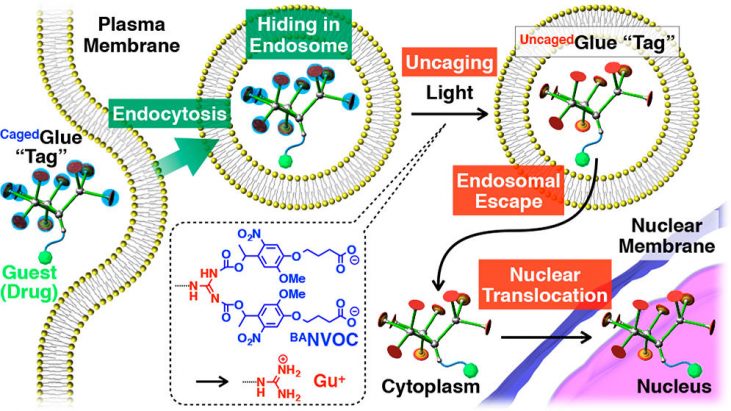
The cell nucleus is one of the most important organelles as a subcellular drug-delivery target. [1] For nucleus-targeted drug delivery, drugs must be designed to overcome several cellular barriers such as the plasma, endosomal, and nuclear membranes. A promising strategy to achieve efficient nuclear translocation of drugs is to attach guanidinium ion (Gu+)-rich tags, because Gu+-rich molecules are known to bind and activate nuclear transporter proteins. T. Aita group at University of Tokyo report caged molecular glues, named Caged Glue-R (Figure 1), which is targeted to cell nucleus and can be site-selectively activated by photo-irradiation. They designed CagedGlue-R by incorporate their previously develoved Guanine dendrimer motif [2] with anionic photocleavable group [3] so that it does not to interact with cell membranes. As illustrated in Figure 2, CagedGlue-R can enter living cells via endocytosis and hide in endosome before it is converted to UncagedGlue-R by photo-irradiation. Subsequently, they are possibly translocated into the nucleus by the action of the Gu+-rich UncagedGlue-R tag.
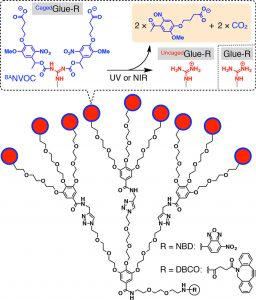
Figure 1 shows the schematic structures of CagedGlue-R, caged molecular glue that bears 9 protected guanidinium ion (Gu+) pendants by a butyrate-substituted nitroveratryloxycarbonyl (BANVOC) group; UncagedGlue-R, photouncaged molecular glue that bears 9 Gu+ pendants; and Glue-R, molecular glues carrying no cage. The focal core of Glue-R is functionalized with either nitrobenzoxadiazole (NBD) or dibenzocyclooctyne (DBCO).
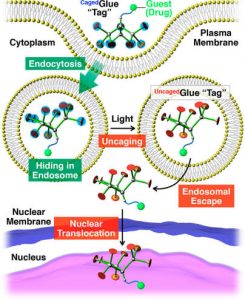
Figure 2 shows the schematic illustration of light-triggered nuclear translocation of guests (drugs) conjugated with a CagedGlue tag. The guest/CagedGlue conjugate is taken up into living cells via endocytosis. Upon photoirradiation, the CagedGlue tag is uncaged to yield an UncagedGlue tag, which strongly interacts with the endosomal membrane and therefore facilitates endosomal escape of the tagged guest. Subsequently, the tagged guest is delivered into the nucleus.
Details
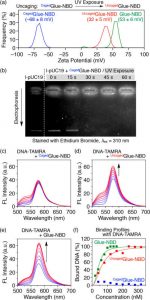
Figure 3 (a) shows zeta potential profiles at 25 °C of CagedGlue-NBD (50 μM) in Tris-HCl (20 mM, pH 7.2) buffer before (blue) and after UV exposure at 365 nm for 60 min (UncagedGlue-NBD, red) with Glue-NBD (50 μM, green) as a reference. (b) Agarose gel electrophoresis profiles (λex = 310 nm) of l-pUC19 in the absence and presence of CagedGlue-NBD before and after UV exposure, with ethidium bromide stain. (c–e) Fluorescence spectra (λex = 480 nm) of DNA-TAMRA (100 nM) at upon titration with (c) CagedGlue-NBD (0–160 nM), (d) UncagedGlue-NBD (0–320 nM), and (e) Glue-NBD (0–160 nM) as a reference. (f) Binding profiles of CagedGlue-NBD (0–320 nM, blue), UncagedGlue-NBD (0–320 nM, red), and Glue-NBD (0–160 nM, green) as a reference to DNA-TAMRA (0.06 nM).
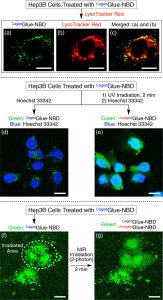
Figure 4 shows confocal laser scanning micrographs of Hep3B cells containing CagedGlue-NBD (10 μM). (a, b) Micrographs upon excitation at (a) 488 nm (λobs = 500–530 nm) and (b) 543 nm (λobs = 565–620 nm) containing LysoTracker Red. (c) Merged image of (a) and (b). (d, e) Micrographs recorded upon excitation at 488 nm (λobs = 500–530 nm, green) and 710 nm (two-photon; λobs = 390–465 nm, blue). The Hep3B cells, treated with CagedGlue-NBD, were incubated at 37 °C in EMEM containing Hoechst 33342 before (d) and after (e) 2 min UV exposure at 365 nm. (f, g) Micrographs recorded upon excitation at 488 nm (λobs = 500–530 nm) before (f) and after (g) two-photon irradiation at 710 nm. The white dashed circle in (f) represents the irradiated area. Scale bars = 20 μm.
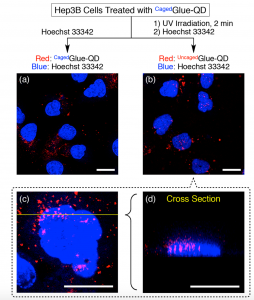
Figure 5 shows confocal laser scanning micrographs of Hep3B cells upon excitation at 405 nm (λobs = 430–520 nm, blue) and 488 nm (λobs = 625–680 nm, red). Hep3B cells were incubated at 37 °C for 3 h in EMEM containing CagedGlue-QD (10 nM), rinsed with D-PBS, and then incubated at 37 °C for 10 min in EMEM containing Hoechst 33342 before (a) and after (b, c) 2 min exposure to UV light at 365 nm. (d) Cross-sectional image along the yellow line in (c). Scale bars = 20 μm.
Reference
[1]”TSubcellular Targeting Strategies for Drug Design and Delivery”
Rajendran, L.; Knölker, H.-J.; Simons, K. Nat. Rev. Drug Discovery 2010, 9, 29−42.
DOI: 10.1038/nrd2897
[2]”Molecular Glues Carrying Multiple Guanidinium Ion Pendants via an Oligoether Spacer: Stabilization of Microtubules against Depolymerization ”
Okuro, K.; Kinbara, K.; Tsumoto, K.; Ishii, N.; Aida, T. J. Am. Chem. Soc. 2009, 131, 1626−1627.
DOI: 10.1021/ja800491v
[3]”Wavelength-Selective Cleavage of Photoprotecting Groups: Strategies and Applications in Dynamic Systems”
Hansen, M. J.; Velema, W. A.; Lerch, M. M.; Szymanski, W.; Feringa, B. L. Chem. Soc. Rev. 2015, 44, 3358−3377.