Yuichiro Hori, Norimichi Otomura, Ayuko Nishida, Miyako Nishiura, Maho Umeno, Isao Suetake, Kazuya Kikuchi. J. Am. Chem. Soc. 140, 5, 1686-1690.
DOI: http://10.1021/jacs.7b09713
ABSTRACT
Hybrid probes consisting of synthetic molecules and proteins are powerful tools for detecting biological molecules and signals in living cells. To date, most targets of the hybrid probes have been limited to pH and small analytes. Although biomacromolecules are essential to the physiological function of cells, the hybrid-probe-based approach has been scarcely employed for live-cell detection of biomacromolecules. Here, we developed a hybrid probe with a chemical switch for live-cell imaging of methylated DNA, an important macromolecule in the repression of gene expression. Using a protein labeling technique, we created a hybrid probe containing a DNA-binding fluorogen and a methylated-DNA-binding domain. The hybrid probe enhanced fluorescence intensity upon binding to methylated DNA and successfully monitored methylated DNA during mitosis. The hybrid probe offers notable advantages absent from probes based on small molecules or fluorescent proteins and is useful for live-cell analyses of epigenetic phenomena and diseases related to DNA methylation.
Introduction
The detecting of various biological molecules and signals in living cells using synthetic-molecule/protein probes have evolved as an advantages method in current bio-imaging. [1] Although biomacromolecules are essential for cell homeostasis and function, fewer studies of hybrid probes for live-cell detection of biomacromolecules have been reported.
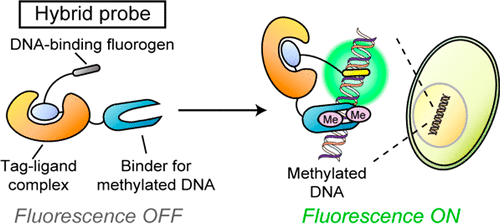
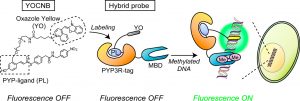
In this paper, Prof. K. Kikuchi and their coworkers developed a hybrid probe for imaging of biomacromolecules, focusing on epigenetically modified DNA. For fluorogenic detection of methylated DNA, they designed a hybrid probe consisting of a DNA-binding fluorogen and PYP3R-tagged MBD1−75 (PYP3R-MBD1−75). Based on their previous work, [2] they first genetically expressed MBD1−75 derived from MBD1 for a module of specific recognizing methylated DNA. For a fluorogen to detect methylated DNA, they designed PYP-ligand-conjugated Oxazole Yellow [3] (YOCNB) and generated the hybrid probe in living cells. (Figure 1)
Figure 1 shows the principle of fluorogenic detection of methylated DNA using a hybrid probe created by a PYP-tag labeling system.
Details
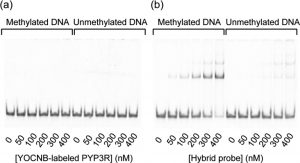
Figure 2 shows the selective binding of a hybrid probe to methylated DNA. The hybrid probe represents the complex of PYP3R-MBD1–75 and YOCNB. After reaction of (a) PYP3R (5 μM) or (b) PYP3R-MBD1–75 (5 μM) with YOCNB (5 μM), the complexes (0, 50, 100, 200, 300, and 400 nM) were incubated with methylated or unmethylated DNA (50 nM) and then analyzed by a gel shift assay.
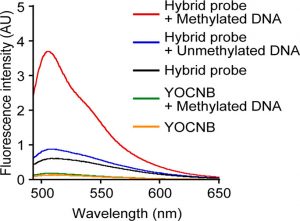 Figure 3 shows the fluorogenic detection of methylated DNA using a hybrid probe. Fluorescence spectra of the hybrid probe (400 nM) and YOCNB (400 nM) in the absence or presence of DNA (50 nM), which was methylated or unmethylated.
Figure 3 shows the fluorogenic detection of methylated DNA using a hybrid probe. Fluorescence spectra of the hybrid probe (400 nM) and YOCNB (400 nM) in the absence or presence of DNA (50 nM), which was methylated or unmethylated.
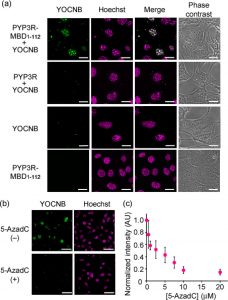
Figure 4 shows the live-cell imaging of methylated DNA. (a) Fluorescence images of cells transfected with empty plasmid and plasmid encoding PYP3R-MBD1–112 or PYP3R. YOCNB (2 μM), Hoechst 33342 (300 ng/mL), and MitoTracker Deep Red (2 μM) were added to the cells. Fluorescent colors of the hybrid probe, Hoechst 33342, and their colocalization are shown as green, magenta, and white in the images, respectively. Scale bar, 20 μm. (b) Effect of 5-AzadC on fluorescence in nuclei. PYP3R-MBD1–112-expressing cells were pretreated in the absence or presence of 5-AzadC (25 μM) for 24 h and then incubated with YOCNB (2 μM) for 75 min. Scale bar, 50 μm. (c) Imaging-based quantification of methylated DNA. Cells expressing PYP3R-MBD1–112 were incubated with different concentrations of 5-AzadC for 24 h and were then labeled with YOCNB (2 μM). The normalized fluorescence intensity of YOCNB in nuclei was plotted against the 5-AzadC concentrations.
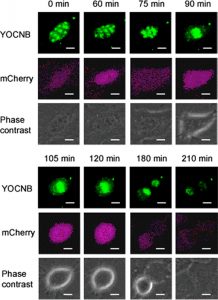
Figure 5 shows the time-lapse imaging of methylated DNA during mitosis. Cells stably expressing mCherry-hGem1–110 were transfected with a PYP3R-MBD1–112-encoding plasmid and were incubated with YOCNB (2 μM) for 40 min. The time-lapse images were obtained using fluorescence microscopy. Scale bar, 5 μm.
Reference
[1]”Recent Progress in Design of Protein-Based Fluorescent Biosensors and Their Cellular Applications”
Tamura, T.; Hamachi, I. ACS Chem. Biol. 2014, 9, 2708−2717.
DOI: 10.1021/cb500661v
[2]”Development of Fluorogenic Probes for Quick No-Wash Live-Cell Imaging of Intracellular Proteins”
Hori, Y.; Norinobu, T.; Sato, M.; Arita, K.; Shirakawa, M.; Kikuchi, K. J. Am. Chem. Soc. 2013, 135, 12360−12365.
DOI: 10.1021/ja405745v
[3]”Synthesis, Photophysical Effects, and DNA Targeting Properties of Oxazole Yellow−Peptide Bioconjugates”
Thompson, M. Bioconjugate Chem. 2006, 17, 507−513.
DOI: 10.1021/bc050307t